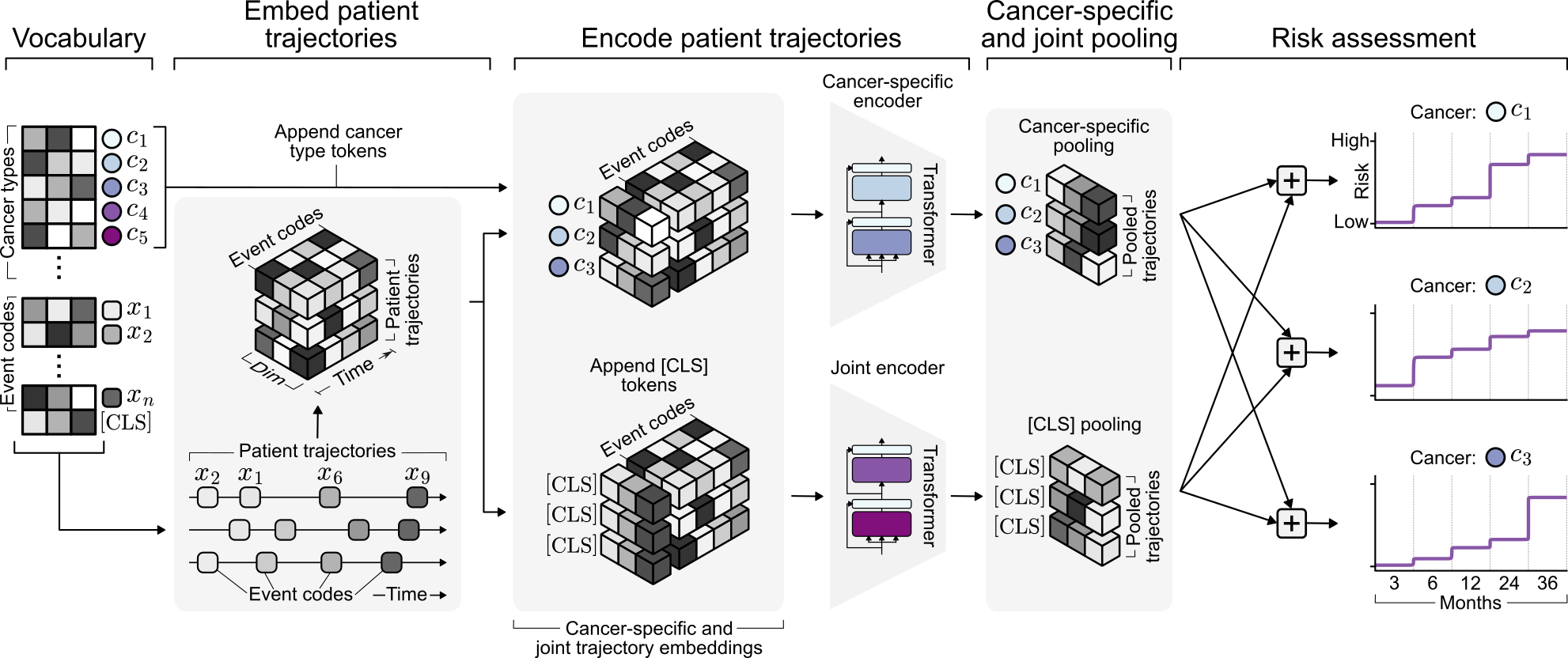
Representation learning from large-scale real-world EHR data
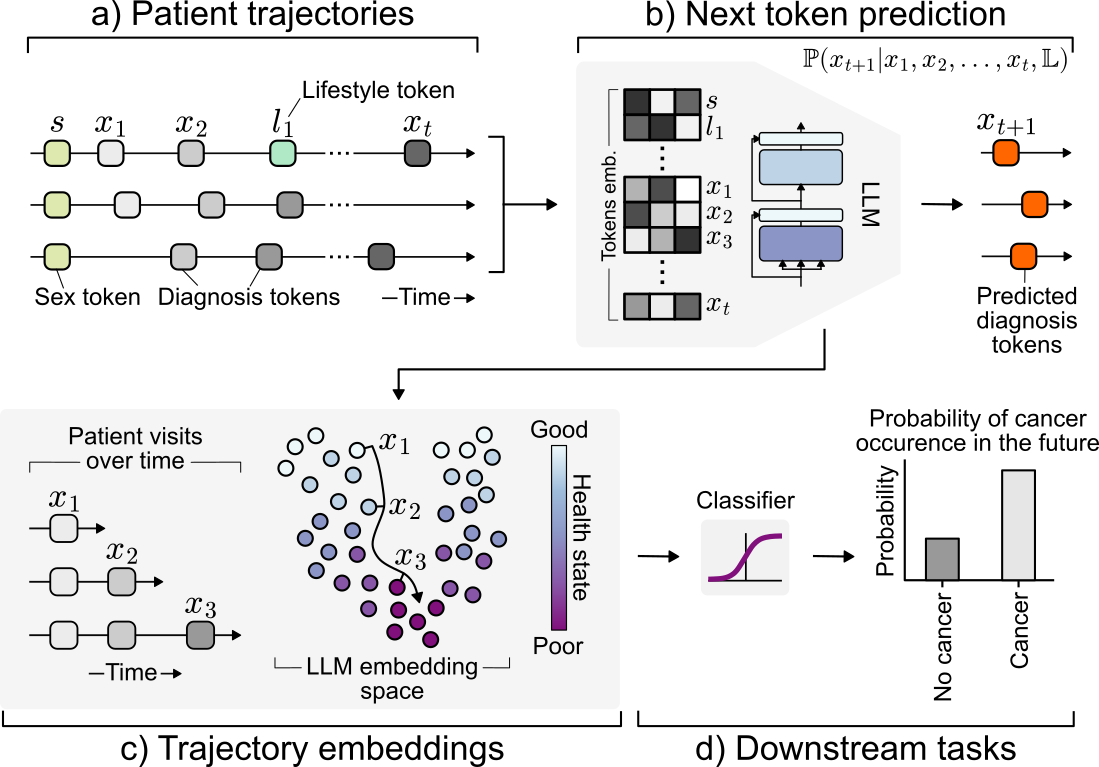
Transforming longitudinal patient trajectories into structured embeddings that capture the evolving health state of patients and support downstream clinical decision-making tasks.